The essentials of R
Manipulation of vector
library(tidyverse)
library(dplyr)
vec <- c(3,5,2,1,5,"O",NA)
length(unique(vec))
## [1] 6
num_vec <- as.numeric(vec)
log(num_vec)
## [1] 1.0986123 1.6094379 0.6931472 0.0000000 1.6094379 NA NA
sum(c(num_vec, NA), na.rm=T)
## [1] 16
sort(num_vec, decreasing = T)
## [1] 5 5 3 2 1
## [1] FALSE FALSE FALSE FALSE FALSE TRUE TRUE
## [1] 3 5 2 1 5
## [1] TRUE FALSE
## [1] FALSE TRUE FALSE FALSE TRUE FALSE FALSE
Generate sequence or repeted sequece
seq(from = 0, to = 10, by = 0.5)
## [1] 0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5 6.0 6.5 7.0
## [16] 7.5 8.0 8.5 9.0 9.5 10.0
## [1] 1 2 3 1 2 3 1 2 3 1 2 3
## [1] 1 1 1 1 2 2 2 2 3 3 3 3
Get directory and write data out and in
## [1] "C:/Users/hed2/Downloads/mybook2/mybook2"
setwd(getwd())
write.csv(cars, "cars.csv", row.names=F)
dataframe <- read.csv("cars.csv")
Function
my_func <- function(x){
x_mod <- (x + 7) * 4
return(x_mod)
}
my_func(num_vec)
## [1] 40 48 36 32 48 NA NA
Plot
plot(dist ~ speed, data=cars)
 ### Build model and plot
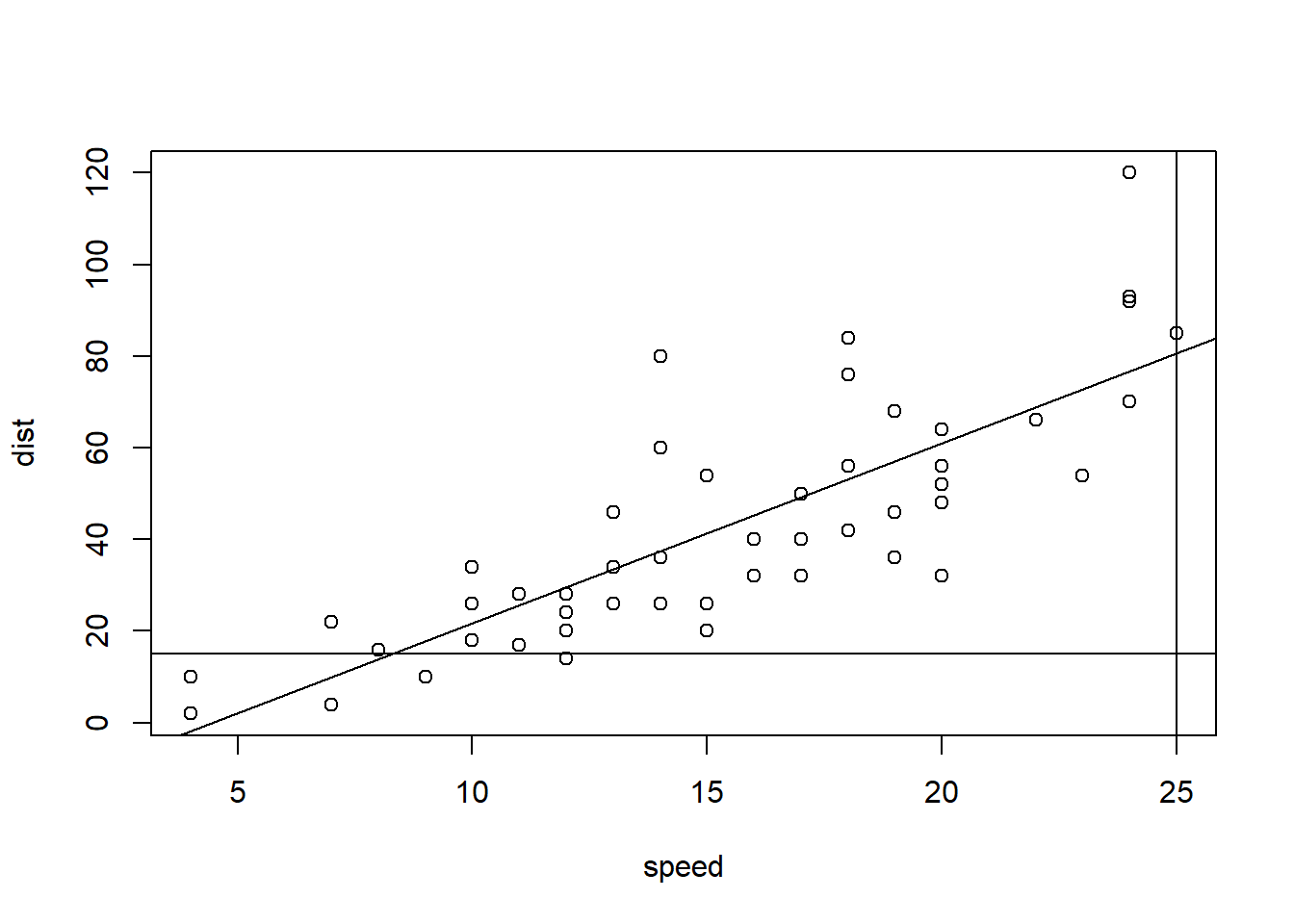
### Build model and plot
model <- lm(dist ~ speed, data=cars)
plot(dist ~ speed, data=cars)
abline(model)
abline(v = 25)
abline(h = 15)
 ### Rename names of columns
### Rename names of columns
## [1] "speed" "dist"
names(cars) <- c("speed per hour", "total dist")
Class of dataframe
matrix <- as.matrix(cars)
df <- as.data.frame(matrix)
class(matrix)
## [1] "matrix" "array"
## [1] "data.frame"
| speed per hour |
4 |
4 |
7 |
7 |
8 |
9 |
10 |
10 |
10 |
11 |
11 |
12 |
12 |
12 |
12 |
13 |
13 |
13 |
13 |
14 |
14 |
14 |
14 |
15 |
15 |
15 |
16 |
16 |
17 |
17 |
17 |
18 |
18 |
18 |
18 |
19 |
19 |
19 |
20 |
20 |
20 |
20 |
20 |
22 |
23 |
24 |
24 |
24 |
24 |
25 |
| total dist |
2 |
10 |
4 |
22 |
16 |
10 |
18 |
26 |
34 |
17 |
28 |
14 |
20 |
24 |
28 |
26 |
34 |
34 |
46 |
26 |
36 |
60 |
80 |
20 |
26 |
54 |
32 |
40 |
32 |
40 |
50 |
42 |
56 |
76 |
84 |
36 |
46 |
68 |
32 |
48 |
52 |
56 |
64 |
66 |
54 |
70 |
92 |
93 |
120 |
85 |
Generate new variable for dataframe (character)
## [1] "raster_1" "raster_2" "raster_3" "raster_4" "raster_5" "raster_6"
## [7] "raster_7" "raster_8" "raster_9" "raster_10"
paste0("raster_", rep(x = 1:5, times = 10))
## [1] "raster_1" "raster_2" "raster_3" "raster_4" "raster_5" "raster_1"
## [7] "raster_2" "raster_3" "raster_4" "raster_5" "raster_1" "raster_2"
## [13] "raster_3" "raster_4" "raster_5" "raster_1" "raster_2" "raster_3"
## [19] "raster_4" "raster_5" "raster_1" "raster_2" "raster_3" "raster_4"
## [25] "raster_5" "raster_1" "raster_2" "raster_3" "raster_4" "raster_5"
## [31] "raster_1" "raster_2" "raster_3" "raster_4" "raster_5" "raster_1"
## [37] "raster_2" "raster_3" "raster_4" "raster_5" "raster_1" "raster_2"
## [43] "raster_3" "raster_4" "raster_5" "raster_1" "raster_2" "raster_3"
## [49] "raster_4" "raster_5"
df$group <- paste0("raster_", rep(x = 1:5, times = 10))
df$id <- paste0("raster_", 1:50)
Create a new dataframe using ‘rnorm’ - random number from distribution
sample <- round((rnorm(50,0, 1)),2)
group <- paste0("raster_", rep(x = 1:5, times = 10))
df_join <- data.frame(sample, group)
df_join$id <- paste0("raster_", 1:50)
Left join two dataframes
library(dplyr)
data_all <- left_join(df, df_join, by="id")
head(data_all)
| 4 |
2 |
raster_1 |
raster_1 |
-0.01 |
raster_1 |
| 4 |
10 |
raster_2 |
raster_2 |
-1.78 |
raster_2 |
| 7 |
4 |
raster_3 |
raster_3 |
-0.78 |
raster_3 |
| 7 |
22 |
raster_4 |
raster_4 |
0.13 |
raster_4 |
| 8 |
16 |
raster_5 |
raster_5 |
-0.71 |
raster_5 |
| 9 |
10 |
raster_1 |
raster_6 |
-0.04 |
raster_1 |
Select variables
select(data_all, group.x, id )
| raster_1 |
raster_1 |
| raster_2 |
raster_2 |
| raster_3 |
raster_3 |
| raster_4 |
raster_4 |
| raster_5 |
raster_5 |
| raster_1 |
raster_6 |
| raster_2 |
raster_7 |
| raster_3 |
raster_8 |
| raster_4 |
raster_9 |
| raster_5 |
raster_10 |
| raster_1 |
raster_11 |
| raster_2 |
raster_12 |
| raster_3 |
raster_13 |
| raster_4 |
raster_14 |
| raster_5 |
raster_15 |
| raster_1 |
raster_16 |
| raster_2 |
raster_17 |
| raster_3 |
raster_18 |
| raster_4 |
raster_19 |
| raster_5 |
raster_20 |
| raster_1 |
raster_21 |
| raster_2 |
raster_22 |
| raster_3 |
raster_23 |
| raster_4 |
raster_24 |
| raster_5 |
raster_25 |
| raster_1 |
raster_26 |
| raster_2 |
raster_27 |
| raster_3 |
raster_28 |
| raster_4 |
raster_29 |
| raster_5 |
raster_30 |
| raster_1 |
raster_31 |
| raster_2 |
raster_32 |
| raster_3 |
raster_33 |
| raster_4 |
raster_34 |
| raster_5 |
raster_35 |
| raster_1 |
raster_36 |
| raster_2 |
raster_37 |
| raster_3 |
raster_38 |
| raster_4 |
raster_39 |
| raster_5 |
raster_40 |
| raster_1 |
raster_41 |
| raster_2 |
raster_42 |
| raster_3 |
raster_43 |
| raster_4 |
raster_44 |
| raster_5 |
raster_45 |
| raster_1 |
raster_46 |
| raster_2 |
raster_47 |
| raster_3 |
raster_48 |
| raster_4 |
raster_49 |
| raster_5 |
raster_50 |
Filter observations
raster_1 <- filter(data_all, group.x == "raster_1")
raster_1
| 4 |
2 |
raster_1 |
raster_1 |
-0.01 |
raster_1 |
| 9 |
10 |
raster_1 |
raster_6 |
-0.04 |
raster_1 |
| 11 |
28 |
raster_1 |
raster_11 |
-0.31 |
raster_1 |
| 13 |
26 |
raster_1 |
raster_16 |
2.19 |
raster_1 |
| 14 |
36 |
raster_1 |
raster_21 |
1.45 |
raster_1 |
| 15 |
54 |
raster_1 |
raster_26 |
0.85 |
raster_1 |
| 17 |
50 |
raster_1 |
raster_31 |
1.11 |
raster_1 |
| 19 |
36 |
raster_1 |
raster_36 |
0.27 |
raster_1 |
| 20 |
52 |
raster_1 |
raster_41 |
-1.49 |
raster_1 |
| 24 |
70 |
raster_1 |
raster_46 |
-2.16 |
raster_1 |
speed_dist <- filter(data_all, data_all$`speed per hour` < 11 & data_all$`total dist` >= 10)
speed_dist
| 4 |
10 |
raster_2 |
raster_2 |
-1.78 |
raster_2 |
| 7 |
22 |
raster_4 |
raster_4 |
0.13 |
raster_4 |
| 8 |
16 |
raster_5 |
raster_5 |
-0.71 |
raster_5 |
| 9 |
10 |
raster_1 |
raster_6 |
-0.04 |
raster_1 |
| 10 |
18 |
raster_2 |
raster_7 |
-0.47 |
raster_2 |
| 10 |
26 |
raster_3 |
raster_8 |
0.61 |
raster_3 |
| 10 |
34 |
raster_4 |
raster_9 |
1.17 |
raster_4 |
Append rows
rbind(raster_1,speed_dist)
| 4 |
2 |
raster_1 |
raster_1 |
-0.01 |
raster_1 |
| 9 |
10 |
raster_1 |
raster_6 |
-0.04 |
raster_1 |
| 11 |
28 |
raster_1 |
raster_11 |
-0.31 |
raster_1 |
| 13 |
26 |
raster_1 |
raster_16 |
2.19 |
raster_1 |
| 14 |
36 |
raster_1 |
raster_21 |
1.45 |
raster_1 |
| 15 |
54 |
raster_1 |
raster_26 |
0.85 |
raster_1 |
| 17 |
50 |
raster_1 |
raster_31 |
1.11 |
raster_1 |
| 19 |
36 |
raster_1 |
raster_36 |
0.27 |
raster_1 |
| 20 |
52 |
raster_1 |
raster_41 |
-1.49 |
raster_1 |
| 24 |
70 |
raster_1 |
raster_46 |
-2.16 |
raster_1 |
| 4 |
10 |
raster_2 |
raster_2 |
-1.78 |
raster_2 |
| 7 |
22 |
raster_4 |
raster_4 |
0.13 |
raster_4 |
| 8 |
16 |
raster_5 |
raster_5 |
-0.71 |
raster_5 |
| 9 |
10 |
raster_1 |
raster_6 |
-0.04 |
raster_1 |
| 10 |
18 |
raster_2 |
raster_7 |
-0.47 |
raster_2 |
| 10 |
26 |
raster_3 |
raster_8 |
0.61 |
raster_3 |
| 10 |
34 |
raster_4 |
raster_9 |
1.17 |
raster_4 |
Create new variables instead of old variables
mutate(data_all,
sample = round(sample,1))
| 4 |
2 |
raster_1 |
raster_1 |
0.0 |
raster_1 |
| 4 |
10 |
raster_2 |
raster_2 |
-1.8 |
raster_2 |
| 7 |
4 |
raster_3 |
raster_3 |
-0.8 |
raster_3 |
| 7 |
22 |
raster_4 |
raster_4 |
0.1 |
raster_4 |
| 8 |
16 |
raster_5 |
raster_5 |
-0.7 |
raster_5 |
| 9 |
10 |
raster_1 |
raster_6 |
0.0 |
raster_1 |
| 10 |
18 |
raster_2 |
raster_7 |
-0.5 |
raster_2 |
| 10 |
26 |
raster_3 |
raster_8 |
0.6 |
raster_3 |
| 10 |
34 |
raster_4 |
raster_9 |
1.2 |
raster_4 |
| 11 |
17 |
raster_5 |
raster_10 |
-0.8 |
raster_5 |
| 11 |
28 |
raster_1 |
raster_11 |
-0.3 |
raster_1 |
| 12 |
14 |
raster_2 |
raster_12 |
1.4 |
raster_2 |
| 12 |
20 |
raster_3 |
raster_13 |
-2.2 |
raster_3 |
| 12 |
24 |
raster_4 |
raster_14 |
-0.3 |
raster_4 |
| 12 |
28 |
raster_5 |
raster_15 |
2.1 |
raster_5 |
| 13 |
26 |
raster_1 |
raster_16 |
2.2 |
raster_1 |
| 13 |
34 |
raster_2 |
raster_17 |
0.2 |
raster_2 |
| 13 |
34 |
raster_3 |
raster_18 |
-0.9 |
raster_3 |
| 13 |
46 |
raster_4 |
raster_19 |
0.2 |
raster_4 |
| 14 |
26 |
raster_5 |
raster_20 |
-0.7 |
raster_5 |
| 14 |
36 |
raster_1 |
raster_21 |
1.4 |
raster_1 |
| 14 |
60 |
raster_2 |
raster_22 |
-0.8 |
raster_2 |
| 14 |
80 |
raster_3 |
raster_23 |
0.4 |
raster_3 |
| 15 |
20 |
raster_4 |
raster_24 |
0.6 |
raster_4 |
| 15 |
26 |
raster_5 |
raster_25 |
-0.8 |
raster_5 |
| 15 |
54 |
raster_1 |
raster_26 |
0.8 |
raster_1 |
| 16 |
32 |
raster_2 |
raster_27 |
-0.8 |
raster_2 |
| 16 |
40 |
raster_3 |
raster_28 |
0.6 |
raster_3 |
| 17 |
32 |
raster_4 |
raster_29 |
1.1 |
raster_4 |
| 17 |
40 |
raster_5 |
raster_30 |
-1.0 |
raster_5 |
| 17 |
50 |
raster_1 |
raster_31 |
1.1 |
raster_1 |
| 18 |
42 |
raster_2 |
raster_32 |
-0.5 |
raster_2 |
| 18 |
56 |
raster_3 |
raster_33 |
0.3 |
raster_3 |
| 18 |
76 |
raster_4 |
raster_34 |
0.2 |
raster_4 |
| 18 |
84 |
raster_5 |
raster_35 |
-0.4 |
raster_5 |
| 19 |
36 |
raster_1 |
raster_36 |
0.3 |
raster_1 |
| 19 |
46 |
raster_2 |
raster_37 |
-1.2 |
raster_2 |
| 19 |
68 |
raster_3 |
raster_38 |
-0.1 |
raster_3 |
| 20 |
32 |
raster_4 |
raster_39 |
0.8 |
raster_4 |
| 20 |
48 |
raster_5 |
raster_40 |
-2.2 |
raster_5 |
| 20 |
52 |
raster_1 |
raster_41 |
-1.5 |
raster_1 |
| 20 |
56 |
raster_2 |
raster_42 |
-1.2 |
raster_2 |
| 20 |
64 |
raster_3 |
raster_43 |
-1.6 |
raster_3 |
| 22 |
66 |
raster_4 |
raster_44 |
0.4 |
raster_4 |
| 23 |
54 |
raster_5 |
raster_45 |
-1.0 |
raster_5 |
| 24 |
70 |
raster_1 |
raster_46 |
-2.2 |
raster_1 |
| 24 |
92 |
raster_2 |
raster_47 |
-0.6 |
raster_2 |
| 24 |
93 |
raster_3 |
raster_48 |
-0.4 |
raster_3 |
| 24 |
120 |
raster_4 |
raster_49 |
0.9 |
raster_4 |
| 25 |
85 |
raster_5 |
raster_50 |
-1.1 |
raster_5 |
summarise statistics
summarise(data_all,
mean_speed = mean(sample),
max_dist = max( "total dist" ))
Group dataframe then summarise statistics
data_all_group <- group_by(data_all, group.x)
summarise(data_all_group,
mean_speed = mean(sample),
max_dist = max( "total dist" ))
Ungroup then summarise statistics
ungroup_data <- ungroup( data_all_group)
summarise( ungroup_data ,
mean_speed = mean(sample),
max_dist = max( "total dist" ))
Summary linear regression model
mod1 <- lm(cars$`total dist` ~ cars$`speed per hour` )
summary(mod1)
##
## Call:
## lm(formula = cars$`total dist` ~ cars$`speed per hour`)
##
## Residuals:
## Min 1Q Median 3Q Max
## -29.069 -9.525 -2.272 9.215 43.201
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -17.5791 6.7584 -2.601 0.0123 *
## cars$`speed per hour` 3.9324 0.4155 9.464 1.49e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 15.38 on 48 degrees of freedom
## Multiple R-squared: 0.6511, Adjusted R-squared: 0.6438
## F-statistic: 89.57 on 1 and 48 DF, p-value: 1.49e-12
Create frequency table
table(data_all_group$`speed per hour`,data_all_group$group.x )
| 4 |
1 |
1 |
0 |
0 |
0 |
| 7 |
0 |
0 |
1 |
1 |
0 |
| 8 |
0 |
0 |
0 |
0 |
1 |
| 9 |
1 |
0 |
0 |
0 |
0 |
| 10 |
0 |
1 |
1 |
1 |
0 |
| 11 |
1 |
0 |
0 |
0 |
1 |
| 12 |
0 |
1 |
1 |
1 |
1 |
| 13 |
1 |
1 |
1 |
1 |
0 |
| 14 |
1 |
1 |
1 |
0 |
1 |
| 15 |
1 |
0 |
0 |
1 |
1 |
| 16 |
0 |
1 |
1 |
0 |
0 |
| 17 |
1 |
0 |
0 |
1 |
1 |
| 18 |
0 |
1 |
1 |
1 |
1 |
| 19 |
1 |
1 |
1 |
0 |
0 |
| 20 |
1 |
1 |
1 |
1 |
1 |
| 22 |
0 |
0 |
0 |
1 |
0 |
| 23 |
0 |
0 |
0 |
0 |
1 |
| 24 |
1 |
1 |
1 |
1 |
0 |
| 25 |
0 |
0 |
0 |
0 |
1 |
Value and variable label
iris$Species <- factor(iris$Species,labels = c( "setosanew","versicolornew","virginianew"))
table(iris$Species)
label(iris$Species) <- "Species types"
table(iris$Species)
Recode a variable
irisifelse <- iris%>%
mutate(Sepal.Length2 = ifelse(Sepal.Length < 6 , "level1", ifelse(Sepal.Length < 7 , "level2", Sepal.Length)))
table(irisifelse$Sepal.Length2)

 ### Build model and plot
### Build model and plot ### Rename names of columns
### Rename names of columns